| front |1 |2 |3 |4 |5 |6 |7 |8 |9 |10 |11 |12 |13 |14 |15 |16 |17 |18 |19 |20 |review |
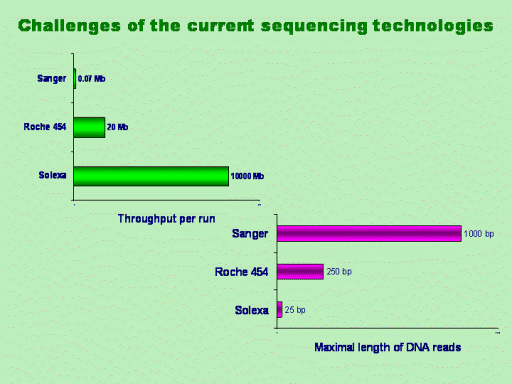 |
Letís consider the general
trends of development of new sequencing technologies. When a
Sanger-based capillary machine that became already a classical
sequencing technique returned approximately 70 kb sequences per a
run with an average length of DNA reads around 800-1000 bp, The
Roche 454 pyro-sequencing technology increased the throughput up to
20 Mb, and the projected throughput of one newly emerging Solexa
technology is as much as 10000 Mb per run. However, the length of
DNA reads is progressively decreasing to 250 and 25 bp respectively.
The expected flood of sequence data, especially from metagenomic
studies, therefore poses many new challenges that urgently need
attention respecting to both: the sheer amount of data and the rich
in complexity.
Now I want present you the
approach to deal with the huge amount of sequence data of the length
around 800 to 1500 bp that corresponds to the classical Sanger-based
sequencing or outputs from Roche 454 after assembling. We have
started a new project addressing the analysis of multiple short DNA
reads of 20-25 bp. This project exploits completely different
approaches. I hope to be able to present in on this meeting next
year.
|